Friday, December 28, 2012
Thursday, December 20, 2012
Monday, December 17, 2012
lit: Tully-Fisher relation in SDSS bands
Sunday, December 16, 2012
Matplotlib: errorbar throws a ValueError: too many values to unpack
To avoid it, check your array dimensions: errorbar(), unlike scatterplot(), keeps on insisting on (x, ) error array dimensions.
1D arrays (vectors) in Numpy can be of shape (x, 1) or (x, ). That's usually interchangeable, but in cases like that, np.reshape(x, (x.shape[0], )) does the trick.
I
Matplotlib: zorder kwarg
scatter_kwargs = {"zorder":100}
error_kwargs = {"lw":.5, "zorder":0}
scatter(X,Y,c=Z,**scatter_kwargs)
errorbar(X,Y,yerr=ERR,fmt=None, marker=None, mew=0,**error_kwargs )
Matplotlib: no edge for marker
Tuesday, December 4, 2012
The select for my TF sample
edit: here's the new one, selecting galaxies with available kinematics data from CALIFA: SELECT distinct h.rowid, n.ba, h.name, j.name, f.sum, n.ba, h.hubtype FROM nadine as n, flags_new as f, morph as h, tf_maps_list as j where h.name =j.name and h.hubtype='S' and h.califa_id = n.califa_id and n.ba > 0.3 and n.ba < 0.9 and f.sum = 0 and f.califa_id = n.califa_id
Pyfits clobber=True keyword
Thursday, November 29, 2012
lit review: Tully-Fisher relation vs. morphology
Monday, November 26, 2012
Scipy: installing from source
some Python scripts from Durham
Sunday, November 25, 2012
awk: making a LaTeX table from a csv file
Converting IDL color table to Python: Matplotlib colour map from rgb array
M. from MPIA wrote a short IDL script that basically takes the rgb distribution vectors across the colour table length, interpolates it to 256 bins and creates a callable colour table.
I thought it would be easy to rewrite it. Matplotlib's documentation was quite incomprehensible. It uses a set of tuples to define a colourmap, which is neat and gives you a lot of control, but I had a different input of colour vectors, which was an array of pre-defined rgb values. colors.ListedColormap did the trick, so here is a script to make a custom colour map from a rgb array with matplotlib.
Friday, November 16, 2012
Seeing structure in the data: histogram bin widths
This problem was sitting at the back of my head for quite some time: we are making interesting claims about some property distributions in our sample, but to what extent are our histograms, i.e. data density distribution plots, robust? How can we claim the histograms represent the true structure (trends, peaks, etc) of the data distribution, while the binning is selected more or less arbitrarily? I think the data should determine how it is represented, not our preconceptions of its distribution and variance.
I tried to redo some of our histograms using Knuth's rule and astroML. Knuth's rule is a Bayesian procedure which creates a piecewise-constant model of data density (bluntly, a model of the histogram). Then the algorithm looks for the best balance between the likelihood of this model (which favours the larger number of bins) and the model prior probability, which punishes complexity (thus preferring a lower number of bins).
Conceptually, think of the bias-variance tradeoff: you can fit anything in the world with a polynomial of a sufficiently large degree, but there's big variance in your model, so its predictive power is negligible. Similarly, a flat line gives you zero variance, but is most probably not a correct representation of your data.
The histogram below, left shows our sample's redshift distribution histogram I found in our article draft. The right one was produced using the Knuth's rule. They might look quite similar, but the left one has many spurious peaks, while the larger bumps on the right one show the _real_ structure of our galaxies distribution in redshift space! The peak near 0.025 corresponds to the Coma galaxy cluster, while the peak at roughly 0.013 shows both the local large-scale structure (Hydra, maybe?) _and_ the abundance of smaller galaxies which do make it into our sample at lower redshifts.
Also consider this pair of histograms:
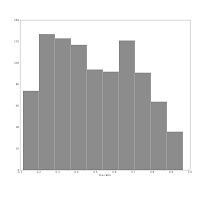
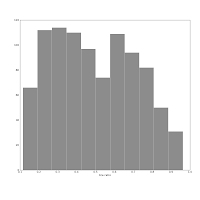 They show the axis ratio (b/a) of our sample of galaxies. The smaller this ratio is, the more inclined we assume the galaxy to be, with some caveats. The left one was produced using matplotlib's default bin number, which is 10, at least in Matlab after which matplotlib is modelled.
They show the axis ratio (b/a) of our sample of galaxies. The smaller this ratio is, the more inclined we assume the galaxy to be, with some caveats. The left one was produced using matplotlib's default bin number, which is 10, at least in Matlab after which matplotlib is modelled.The right one shows the histogram produced using Knuth's rule. It shows the real data distribution structure: the downward trend starting at the left shows that we have more inclined, disk galaxies in our sample (which is true). The bump at the right, at b/a = 0.7, is the population of rounder elliptical galaxies. The superposition of these two populations is shown much more clearly in the second histogram, and we can make some statistical claims about it, instead of just trying to find a pattern and evaluate it visually. Which is good, because we the humans tend to find patterns everywhere and the noise in astronomical datasets is usually large.
Thursday, November 15, 2012
location of Python module file
Friday, November 9, 2012
Tuesday, November 6, 2012
ds9 to NumPy pixel coordinates
I'll have to update the galaxy center pixel coordinates, as the WCS information is taken from the original SDSS images. They would not have to change if the cropped part is below and right the original center.
I don't have a paper and a pencil at the moment, so I'm thinking out loud here. NumPy indexing goes like this, y first:
+------------------> x | | | | V y
ds9, however, uses the standard mathematical notation: (x, y), where x goes left to right, and y goes up from the lower left corner:
y A | | | | + -------------------------> x
This array had its (0, 0) and (10, 20) pixels set to 1:
[[ 1. 0. 0. ..., 0. 0. 0.] [ 0. 0. 0. ..., 0. 0. 0.] [ 0. 0. 0. ..., 0. 0. 0.] ..., [ 0. 0. 0. ..., 0. 0. 0.] [ 0. 0. 0. ..., 0. 0. 0.] [ 0. 0. 0. ..., 0. 0. 0.]]And here is the corresponding ds9 view: tl;dr: if you get a pair of pixel coordinates from ds9 and want to use them in NumPy, you have to swap the coordinate numbers and flip the y-axis, i.e. subtract the ds9 y coordinate from the total image height (image.shape[0]).
Matplotlib: XKCDify, bimodal Gaussians
Monday, November 5, 2012
Matplotlib: intensity map with hexbin
Thursday, November 1, 2012
SQLite: one more query
Thursday, October 25, 2012
Histogram widths: Bayesian blocks
The utility of the Bayesian blocks approach goes beyond simple data representation, however: the bins can be shown to be optimal in a quantitative sense, meaning that the histogram becomes a powerful statistical measure.
Wednesday, October 24, 2012
Photometry: linear regression fitting of sky
The idea I had this morning on the train was simple -- I don't know what conceptual block prevented me from going this way sooner.
Basically, having a cumulative flux curve (the growth curve) is not generally helpful, as the growth of flux outside is non-linear (depends on geometry as well as on the sky value). However, if I normalise the flux profile to _flux_per_pixel, it should theoretically flatten far away from the galaxy. The slope of a linear regression fit should show the quality of the sky value -- the level of variations. If the sky slope is negative and pretty large, then we probably still are within the galaxy.
If the slope is reasonably small (here go the arbitrary parameters again..), simply taking a mean of all measurements within the ring would give a reasonably good sky value.
The catch is getting the width of the elliptical ring used for fitting right. (I can get its distance by waving my hands, taking the maximum distance from my previous measurements, multiplying it by pi/2 or something. We're testing for it anyway).
However, the width of this ring is a tradeoff between accuracy and precision. Taking a wider ring would surely help to reduce the scatter due to random noise, arbitrary gradients and so. However, the possibility to get a (poorly) masked region or some sky artifact, etc. inside this ring also increases.
I tested it a bit using scipy.linalg routines, so far the slope was below 10^-4 counts.
The growth curve itself is useful as a sky subtraction quality check.
Monday, October 22, 2012
awk one-liner
awk 'BEGIN {FS =","}; {print $1, $6, $7, $8, $9}' catalogue.csv > cat.csv
Thursday, October 18, 2012
SQLite: some average values
select z.califa_id, z.z_mag - m.petroMag_z, z.z_mag, m.petroMag_z from z_test as z, mothersample as m where m.califa_id = z.califa_id
Monday, October 15, 2012
SQLITE: query for multiple quantities from several tables
SELECT g.califa_id, g.r_mag, g.el_hlma, l.hlr, m.isoA_r, g.ba, g.flags, r.mean_sky,
r.gc_sky FROM gc as g, gc_r as r, lucie as l, mothersample as m where (g.califa_id
= r.califa_id) and (g.califa_id = m.califa_id) and (g.califa_id = l.califa_id)
and (g.el_hlma > 25) order by g.el_hlma desc
Friday, October 12, 2012
mailx: email a file from shell
Thursday, October 11, 2012
orientation angles of SDSS images wrt North
A wrapper for wrapper for kcorrect
SQLITE: useful query: matching two tables by ra, dec
SELECT m.CALIFA_ID, round(m.ra, 5), round(m.dec, 5), round(s.ra, 5), round(s.dec, 5)
from mothersample as m, sdss_match as s where (round(m.ra, 5) = round(s.ra, 5)
and round(m.dec, 5) = round(s.dec, 5)) order by CALIFA_ID asc
SELECT m.rowid, m.CALIFA_ID, round(m.ra, 5), round(m.dec, 5), s.petroMagErr_u, s.petroMagErr_g, s.petroMagErr_r, s.petroMagErr_i, s.petroMagErr_z, s.extinction_u, s.extinction_g, s.extinction_r, s.extinction_i, s.extinction_z from mothersample as m, sdss_match as s where (round(m.ra, 5) = round(s.ra, 5) and round(m.dec, 5) = round(s.dec, 5)) order by m.rowid
NED positional query -- a script to export ra, dec from database
Wednesday, October 10, 2012
SQLite: field names and leading space
sqlite3.OperationalError: no such column: el_hlma. The column name had a leading space before it, that's why the query failed.
Merging two csv files -- replacing some lines with those from the second
Tuesday, October 9, 2012
NumPy: around() and a way around it
Sunday, October 7, 2012
SDSS CASjobs -- looking for objects within a given radius of a coordinate pair
select p.ra,p.dec,p.b,p.l,p.objID,p.run,p.rerun,p.camcol,p.field,p.obj,p.type,p.flags,p.fiberMag_r,p.petroMag_u,p.petroMag_g,p.petroMag_r,p.petroMag_i,p.petroMag_z,
p.petroRad_r,p.petroR50_r,p.petroR90_r,p.isoA_g,p.isoB_g,p.isoA_r,p.isoB_r,p.isoPhi_r,p.specObjID into mydb.MiceB from PhotoObjAll as p,n
WHERE p.objID = n.objID
and
( flags & (dbo.fPhotoFlags('NOPETRO') +
dbo.fPhotoFlags('MANYPETRO')
+dbo.fPhotoFlags('TOO_FEW_GOOD_DETECTIONS')) ) = 0
Wednesday, October 3, 2012
ls -- sorting files by size
Friday, September 28, 2012
SQLITE -- a useful join
Monday, September 24, 2012
Deploying my inpainting code on other machines
required:
wget
python (developed on 2.6.5)
First, I need pyfits:
mkdir python
cd python
wget http://pypi.python.org/packages/source/p/pyfits/pyfits-3.1.tar.gzSecond, pyfits need setuptools:
wget http://pypi.python.org/packages/2.7/s/setuptools/setuptools-0.6c11-py2.7.egg#md5=fe1f997bc722265116870bc7919059ea
I want to install to a custom location (the ./python directory tree), hence I have to create directory trees for both of them:
mkdirhier [ABSOLUTE PATH TO]/python/lib/python2.7/site-packages/
mkdirhier [ABSOLUTE PATH TO]/python/lib64/python2.7/site-packages/
The .bashrc file's PYTHONPATH variable must be adjusted:
vi ~/.bashrc
export PYTHONPATH=[ABSOLUTE PATH TO]/python/lib/python2.7/site-packages:[ABSOLUTE PATH TO]/python/lib64/python2.7/site-packages/Do not forget to restart bash:
bash
Then, installing setuptools:sh setuptools-0.6c11-py2.7.egg --prefix='[PATH TO]/python'
python setup.py install --prefix='[PATH TO]/python'
../growth-curve-photometry/
../data/SDSS/u/ #griz
../data/MASKS/
../data/maskFilenames.csv
../data/maskFilenames.csv
../data/SDSS_photo_match.csv
../data/CALIFA_mother_simple.csv
../data/filled_u #griz
I need AstLib as well:
wget http://downloads.sourceforge.net/project/astlib/astlib/0.6.1/astLib-0.6.1.tar.gz?r=http%3A%2F%2Fastlib.sourceforge.net%2F&ts=1348484024&use_mirror=switch
tar -xvzj astLib-0.6.1.tar.gz
cd astLib
python setup.py install --prefix='[PATH TO]/python/'
...and all the hell breaks loose here, because my home is mounted across the network, hence $PYTHONPATH points to a couple of different Python versions. Separate ~.bashrc.$HOSTNAME files worked: http://askubuntu.com/questions/15618/repercussions-to-sharing-bashrc-across-machines-with-dropbox.
Then fill.py should be edited, or even copied to allow several different processes work in different bands and galaxy sample slices.
Friday, September 7, 2012
making posters with LaTeX, rendering matplotlib images of right size
Some things I learned underway, from matplotlib cookbook mostly:
For an A0 poster, getting image sizes and font sizes right is important. While compiling my .tex document, I inserted
\showthe\linewidth
fig_width_pt = 628.62 # Get this from LaTeX using \showthe\columnwidth
inches_per_pt = 1.0/72.27 # Convert pt to inches
golden_mean = (sqrt(5)-1.0)/2.0 # Aesthetic ratio
fig_width = fig_width_pt*inches_per_pt # width in
fig_height = fig_width*golden_mean+0.3 # height in inches
fig_size = [fig_width,fig_height]
print fig_size
fig_size was later set as a matplotlib parameter. If you specify .pdf as the filename extension, matplotlib generates a crisp file to be used as an image by LaTeX.
Sunday, September 2, 2012
Getting a single file from git stash
A solution from stackoverflow:
git checkout stash@{0} -- [filename]
opening images with ds9 from a python script
import os
os.system("~/ds9 -zoom 0.3 -scale mode 99.5 -file "+
GalaxyParameters.getSDSSUrl(listFile, dataDir, i) +" -file "+
GalaxyParameters.getMaskUrl(listFile, dataDir, simpleFile, i) +" -match frames")
Monday, August 27, 2012
Decent scientific plots with matplotlib
For instance, histogram bar widths: they are different for different datasets, and you cannot compare two distributions if that's the case. You have to resort to hacks like this:
hist, bins = np.histogram(data, bins = 10)
width=1*(bins[1]-bins[0])
So, what makes an ugly plot look decent? First of all, ticks:
minor_locator = plt.MultipleLocator(plotTitles.yMinorTicks)
Ymajor_locator = plt.MultipleLocator(plotTitles.yMajorTicks)
major_locator = plt.MultipleLocator(plotTitles.xMajorTicks)
Xminor_locator = plt.MultipleLocator(plotTitles.xMinorTicks)
ax.xaxis.set_major_locator(major_locator)
ax.xaxis.set_minor_locator(Xminor_locator)
ax.yaxis.set_major_locator(Ymajor_locator)
ax.yaxis.set_minor_locator(minor_locator)
Some parameters, redefining matplotlib's defaults (these plots are for printouts, so font sizes are big):
params = {'backend': 'ps',
'axes.labelsize': 10,
'text.fontsize': 10,
'legend.fontsize': 10,
'xtick.labelsize': 8,
'ytick.labelsize': 8,
'text.usetex': True,
'font': 'serif',
'font.size': 16,
'ylabel.fontsize': 20}
markers = itertools.cycle('.^*')
p1 = ax.plot(gd.data[0], gd.data[1], linestyle='none', marker=markers.next(), markersize=8, color=gd.colour, mec=gd.colour, alpha = 1)
I would not claim the plots shown are publication-quality (just look at slightly differing histogram widths). But they look way better than default plots one would get with matplotlib.
RGB colours in matplotlib plots
\definecolor{AIPblue} {RGB}{0, 65, 122}
#colour settings
ASColour = '#666666'
Forcing capital letters in Bibtex bibliography lists
Saturday, August 25, 2012
Generating matplotlib plots via ssh on a remote server
_tkinter.TclError: no display name and no $DISPLAY environment variable
Friday, August 24, 2012
Failed optimisation: numpy.roll(), masked arrays
Besides, there's really some room for improvement: those loops and exceptions are ugly and probably avoidable, and looping over array many times could be sped up using some numpy kung-fu.
I wrote a prototype that juggles with the masks, uses some boolean manipulations and numpy.roll(). Near the edges the algorithm reverts to traditional loops over indices.
However, it's still a slow. I ran a profiler: the loops shifting the array take 0.5s each, and so I win nothing in the long run. Array operations are expensive, and when an array is indexed repeatedly within a loop, the code gets slow inevitably. But there's no way I can escape recursion, because I have to pick up a new set of pixels every time.
I still have some ideas, but I want to have some time to analyse the results.
Thursday, August 23, 2012
Morning coffee read -- ps
Wednesday, August 22, 2012
The new inpainting script
It maps the number of all finite (not NaNs, not infs, not having indices beyond the array edges) neighbours of each pixel in the given array. Then it loops through the masked pixels which have the _largest_ number of adjacent neighbours available, picks the closest neighbours in a 5*5 matrix around each pixel, fills the masked pixel with distance-weighted Gaussian average of said neighbours, then updates the neighbour count, loops through the pixels which have the largest number (out of 4) of neighbours again, (...).
I realised I should do inpainting this way while riding my bicycle from cbase. My previous implementation simply looped over indices along one axis, disregarding the number of pixels used for filling the given one. Hence ugly flux shape deformations when a masked region is directly above or below the galaxy in question.
But now it seems to be fine! So finally I get to do some science, instead of coding 24 if's and exceptions in one function.
randomize contents of a list
Updated -- no, it didn't. The weird lines and gradients are the edge effects of rectangular mask shapes, which I didn't like from the beginning. On the other hand, given that the inpainted values vary by ~4 counts (the contrast is stretched in this image), I'm not going to worry about this.
Monday, August 20, 2012
SciPy: spatial binary tree
Numpy: getting indices of n-dimensional array
Inpainting, optimisation, nearest neighbours in Numpy
My current idea is to do this recursively: first fill (using the same Gaussian kernel) the pixels that have the nearest
Here's some food for thought: http://stackoverflow.com/questions/9132114/indexing-numpy-array-neighbours-efficiently, http://davescience.wordpress.com/2011/12/12/counting-neighbours-in-a-python-numpy-matrix/, http://stackoverflow.com/questions/5551286/filling-gaps-in-a-numpy-array.
Sunday, August 19, 2012
batch command line image resizing on remote server
Batch conversion of fits files to .png images, dynamic range and histogram equalisation
scipy.misc.imsave('img/output/filled/'+ID+'_imsave.jpg', outputImage)
Friday, August 17, 2012
matplotlib: missing x, y axis labels
plt.ylabel = plotTitles.ylabel
plt.ylabel(plotTitles.ylabel)
Getting a fits header keyword value from a list of files
#a rough excerpt from ellipse_photometry module.
#loops through filenames, extracts fits header keyword values.
sky = np.empty((939, 2))
for i in range(0, 939):
try:
fileName = GalaxyParameters.getSDSSUrl(listFile, dataDir, i)
head = pyfits.open(fileName)[0].header
sky[i:, 0] = i+1
sky[i:, 1] = head['SKY']
print head['SKY']
except IOError as e:
sky[i:, 0] = i+1
sky[i:, 1] = 0
pass
except KeyError as err:
sky[i:, 0] = i+1
sky[i:, 1] = -1
pass
np.savetxt('SDSS_skies.txt', sky, fmt = ['%i', '%10.5f'])
Drawing a diagonal (or any arbitrarily inclined and offset) line in Matplotlib
import pylab as pl
import numpy as np
x1,x2,n,m,b = min(data),max(data).,1000,1.,0.
# 1000 -- a rough guess of the sufficient length of this line.
#you can actually calculate it using the Pythagorean theorem.
#1, here -- the slope, 0 -- the offset.
x = np.r_[x1:x2:n*1j] #http://docs.scipy.org/doc/numpy/reference/generated/numpy.r_.html
pl.plot(x,m*x + b); pl.grid(); pl.show()
Thursday, August 16, 2012
Image intensity scaling
Bulges and ellipticals
He sums up a great deal of recent research regarding the structure of galaxies. A few main points:
- the components that we call bulges are not necessarily old 'scaled down' ellipticals as we often hear: there are classical bulges, disk-like bulges, 'box-peanuts', which may coexist or overlap.
- 'box-peanuts' are thought to be the inner parts of bars, not separate components
- classical bulges are kinematically hot, i.e. supported by their velocity dispersion. disk-like bulges are kinematically cold, i.e. supported by their rotation velocity.
- there's an interesting section on photometric bulge recognition, and why the arbitrary cut on n=2 is what it is, arbitrary. I tend to believe that any classification has to be data-driven, so I'll keep the alternative suggestions in mind.
- another interesting part is about the scaling relations, i.e. the fundamental plane and F-J relation. I didn't know that the FP can be derived directly from the virial theorem!
- F-J and L-size relations seem to be nonlinear, according to the data.
Wednesday, August 15, 2012
Git -- non-fast-forward updates were rejected
To prevent you from losing history, non-fast-forward updates were rejected
Merge the remote changes before pushing again. See the 'Note about
fast-forwards' section of 'git push --help' for details.
Git -- fixing a detached head
I returned from the state of detached head this way.
'In Git commits form a directed acyclic graph where each commit points to one or more parent commits'.
Geek language, you're a thing of beauty.Monday, August 13, 2012
Python: convert a numpy array to list
inputImage = np.zeros(inputImage.shape)
for i in range(0, inputImage.shape[0]):
for j in range(0, inputImage.shape[1]):
inputIndices[i, j] = (i, j)
inputIndices = inputIndices.tolist()
Python: removing duplicate elements from a list
out = [(12, 3), (11, 4), (12, 3), (44, -5)]
out = list(set(out)))
Thursday, August 9, 2012
unix: show sizes of all files and directories, without subdirectories
du -ah --max-depth=1
numpy array slices: inclusivity vs.exclusivity
fluxData[distance-5:distance, 3]
The idea was to average an array over a running 5 element width annulus, counting back from the distance'th element.
I normally know this, but Numpy's array slicing does not include the last element, i.e. distance'th row was excluded from the count.
For instance,
fluxData[distance-4:distance+1, 3] looks stupid, but somehow worked.
Inpainting again
Monday, May 28, 2012
Set values to null in SQLite; table updating
UPDATE Table SET z = z/300000 where abs(z) > 2
Left outer joins with SQLITE
Saturday, May 26, 2012
SDSS axis ratio measures
The model fits yield an estimate of the axis ratio and position angle of each object, but it is useful to have model-independent measures of ellipticity. In the data released here, frames provides two further measures of ellipticity, one based on second moments, the other based on the ellipticity of a particular isophote. The model fits do correctly account for the effect of the seeing, while the methods presented here do not.
The first method measures flux-weighted second moments, defined as:
- Mxx = x^2/r^2
- Myy = y^2/r^2
- Mxy = x^y/r^2
In the case that the object's isophotes are self-similar ellipses, one can show:
- Q = Mxx - Myy = [(a-b)/(a+b)]cos2φ
- U = Mxy = [(a-b)/(a+b)]sin2φ
where a and b are the semi-major and semi-minor axes, and φ is the position angle. Q and U are Q and U in PhotoObj and are referred to as "Stokes parameters." They can be used to reconstruct the axis ratio and position angle, measured relative to row and column of the CCDs. This is equivalent to the normal definition of position angle (East of North), for the scans on the Equator. The performance of the Stokes parameters are not ideal at low S/N. For future data releases, frames will also output variants of the adaptive shape measures used in the weak lensing analysis of Fischer et al. (2000), which are closer to optimal measures of shape for small objects.
Isophotal QuantitiesA second measure of ellipticity is given by measuring the ellipticity of the 25 magnitudes per square arcsecond isophote (in all bands). In detail, frames measures the radius of a particular isophote as a function of angle and Fourier expands this function. It then extracts from the coefficients the centroid (isoRowC,isoColC), major and minor axis (isoA,isoB), position angle (isoPhi), and average radius of the isophote in question (Profile). Placeholders exist in the database for the errors on each of these quantities, but they are not currently calculated. It also reports the derivative of each of these quantities with respect to isophote level, necessary to recompute these quantities if the photometric calibration changes.
Many more about those second order moments, and why they are called 'Stokes' parameters' can be found in references herein.Thursday, May 24, 2012
Including source code in LaTeX document
Counting occurences of a particular word in a file
i had to calculate how many objects NED database found near determined positions had no definite redshifts. I parsed the NED output files using NED_batch_compact_parser.py by Min-Su Shin.
NED writes out None for quantities it does not find, so I used grep -o None ned_output.txt | wc -wWednesday, May 23, 2012
NED batch job form
It's amazing what contrived workarounds are constructed in public astronomical databases in order to avoid letting people see and harness the SQL underneath. Here's a setup file for NED batch jobs tool I made to work finally:
OUTPUT_FILENAME test.csv OUTPUT_OPTION compact COMPRESS_OPTION none INPUT_COORDINATE_SYSTEM equatorial OUTPUT_COORDINATE_SYSTEM equatorial INPUT_EQUINOX J2000.0 OUTPUT_EQUINOX J2000.0 EXTENDED_NAME_SEARCH yes OUTPUT_SORTED_BY Distance_to_search_center REDSHIFT_VELOCITY 1000.0 SEARCH_RADIUS 0.05 BEGIN_YEAR 1900 END_YEAR 2012 IAU_STYLE S FIND_OBJECTS_NEAR_POSITION 177.34806988d; -1.08377011d; 0.05 REDSHIFT Unconstrained UNIT z INCLUDE INCLUDE ALL Galaxies X GClusters X Supernovae _ QSO X AbsLineSys _ GravLens _ Radio _ Infrared _ EmissnLine X UVExcess X Xray X GammaRay _ GPairs _ GTriples _ GGroups _ END_INCLUDE EXCLUDE Galaxies _ GClusters _ Supernovae _ QSO _ AbsLineSys _ GravLens _ Radio _ Infrared _ EmissnLine _ UVExcess _ Xray _ GammaRay _ GPairs _ GTriples _ GGroups _ END_EXCLUDE END_OF_DATA END_OF_REQUESTS
Left outer joins in SDSS
I was selecting two samples from the SDSS DR7: one which was only redshift-selected, and another which had no cuts in redshift, but was diameter-limited. I wanted to keep the redshift information for the diameter-limited sample (wherein not all galaxies have spectroscopic redshifts), if it was available for a particular galaxy. Hence, outer joins.
Tuesday, April 24, 2012
numpy: combining 'less' and 'more' operators in where expression, bitwise AND
Efficiency 101: reading large FITS table with Python
Last week I hastily wrote some code to search the NSAtlas. It was almost ok for a single galaxy: the array is huge, and iterating through it 1000 times...it took 1 minute for each loop, so I would have to wait all day for the full query to complete.
I've changed the 'if' loop to numpy.where statements and used the collections module: here. It is almost usable, though it takes 15 minutes to search for some variable for 1000 galaxies, and the machine slows down to an almost complete halt.
There is a significant overhead associated with opening and closing files, especially as the one in question was quite huge (0.5 GB) in this case. Not calling the function from within a loop, but passing an array of search arguments and then looping through it within the function reduced the running time to 16 seconds. A 60-fold improvement in two minutes.
I'd also like to mention a fine module called collections, which helped me find an overlap between two lists, like that:
a_multiset = collections.Counter(list(ras[0]))
b_multiset = collections.Counter(list(decs[0]))
overlap = list((a_multiset & b_multiset).elements())Wednesday, April 18, 2012
NSAtlas, surface photometry
Monday, April 16, 2012
Surface photometry
Friday, April 13, 2012
SDSS database utilities
Thursday, April 12, 2012
Latex degree symbol
Wednesday, April 11, 2012
shell: counting files in the current directory
Wednesday, April 4, 2012
Creating a discrete Gaussian kernel with Python
def gauss_kern(size, sizey=None):
""" Returns a normalized 2D gauss kernel array for convolutions """
size = int(size)
if not sizey:
sizey = size
else:
sizey = int(sizey)
x, y = mgrid[-size:size+1, -sizey:sizey+1]
g = exp(-(x**2/float(size)+y**2/float(sizey)))
return g / g.sum()
def gauss_kern(size, sizey=None):
""" Returns a normalized 2D gauss kernel array for convolutions """
size = int(size)
if not sizey:
sizey = size
else:
sizey = int(sizey)
x, y = scipy.mgrid[-size:size+1, -sizey:sizey+1]
g = scipy.exp(-(x**2/float(size)+y**2/float(sizey)))
return g / g.max()
Tuesday, April 3, 2012
A quick way to check if a list of files is complete
# -*- coding: utf-8 -*-
import os
import csv
import string
def touch(fname, times = None):
with file(fname, 'a'):
os.utime(fname, times)
for i in range(0, 939):
with open('../data/maskFilenames.csv', 'rb') as f:
mycsv = csv.reader(f)
mycsv = list(mycsv)
fname = string.strip(mycsv[i][1])
touch(fname)
print fname
Check if files in a list exist with Python
wrongFilenames = []
for i in range(0, noOfGalaxies):
maskFile = GalaxyParameters.getMaskUrl(listFile, dataDir, simpleFile, i)
print i, 'maskfile: ', maskFile
try:
f = open(maskFile)
print "maskfile exists:", maskFile
except IOError as e:
wrongFilenames.append(maskFile)
Creating masks with GIMP
masked = np.where(mask != 0)
mask[masked] = mask[masked]/mask[masked]
Monday, April 2, 2012
WCS to pixel values and vice versa in Python
from astLib import astWCS
WCS=astWCS.WCS("Galaxy.fits")
center = WCS.wcs2pix(ra, dec)
print center
print 'brightness at the galaxy center', img[center[1], center[0]]
Getting tsField files from SDSS
wget http://das.sdss.org/imaging/'+run+'/'+rerun+'/calibChunks/'+camcol+'/tsField-'+runstr+'-'+camcol+'-'+rerun+'-'+field_str+'.fit
Friday, March 23, 2012
M. Kramer on pulsars
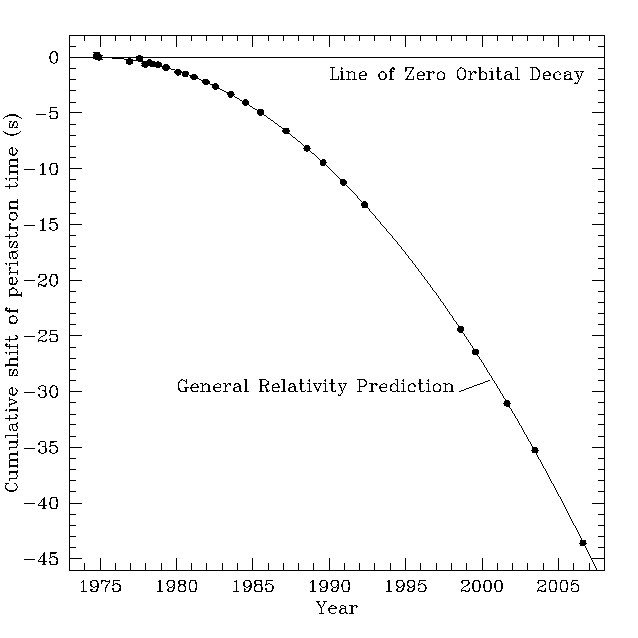
Michael Kramer (MPIfR/JBCA) gave a captivating and eloquent talk this morning on using pulsars as tests for GR and as constraints on alternative theories of gravity. It's possible to use pulsar data to measure post-Keplerian parameters of the only known double pulsar system, and GR passes all the tests with flying colours. Measurements of other binary pulsar systems (PS + a white dwarf, for example) very probably put an end to MOND and other TeVeS gravity theories.
Pulsar timing is precise to 10^-18 s, more precise than atomic clocks and it's apparently possible to notice irregularities in the terrestrial atomic timescale. Perihelion precession of pulsar binary systems is of order of degrees/year, meaning that Newtonian dynamics are completely inadequate here (one would be hopelessly lost soon, if she tried to navigate her spaceship in the system using Newton's dynamics. The stars would be out of place).This image, showing the change in revolution time in binary pulsar system, came up. It's the strongest proof of gravitational wave existence so far, showing how the system radiates its energy away as ripples in the spacetime. The uncertainties are too small to picture.
Interesting results coming up, something to keep an eye on.Thursday, March 22, 2012
SDSS images batch downloading, getting key values from .fits headers
I'm using a couple of string manipulation functions from sdsspy, I felt it's really pointless to rewrite those.
import os
import pyfits
import csv
import numpy
import string
import gzip
#awk < sdss_database_dump.csv 'BEGIN { FS="," }; { print $1,",",$2,",",$3,",",$8,",",$9,",",$10,",",$11}' > list.txt
dataFile = 'list.txt'
#the next three functions are from sdsspy library (http://code.google.com/p/sdsspy/)
def run2string(runs):
"""
rs = run2string(runs)
Return the string version of the run. 756->'000756'
Range checking is applied.
"""
return tostring(runs,0,999999)
def field2string(fields):
"""
fs = field2string(field)
Return the string version of the field. 25->'0025'
Range checking is applied.
"""
return tostring(fields,0,9999)
def tostring(val, nmin=None, nmax=None):
if not numpy.isscalar(val):
return [tostring(v,nmin,nmax) for v in val]
if isinstance(val, (str,unicode)):
nlen = len(str(nmax))
vstr = str(val).zfill(nlen)
return vstr
if nmax is not None:
if val > nmax:
raise ValueError("Number ranges higher than max value of %s\n" % nmax)
if nmax is not None:
nlen = len(str(nmax))
vstr = str(val).zfill(nlen)
else:
vstr = str(val)
return vstr
csvReader = csv.reader(open(dataFile, "rU"), delimiter=',')
f = csv.writer(open('angles.txt', 'w'), delimiter=',')
for row in csvReader:
print '********************************', row[0]
ID = string.strip(row[0])
ra = string.strip(row[1])
dec = string.strip(row[2])
run = string.strip(row[3])
rerun = string.strip(row[4])
camcol = string.strip(row[5])
field = string.strip(row[6])
runstr = run2string(run)
field_str = field2string(field)
print 'wget http://das.sdss.org/imaging/'+run+'/'+rerun+'/corr/'+camcol+'/fpC-'+runstr+'-r'+camcol+'-'+field_str+'.fit.gz'
os.system('wget http://das.sdss.org/imaging/'+run+'/'+rerun+'/corr/'+camcol+'/fpC-'+runstr+'-r'+camcol+'-'+field_str+'.fit.gz')
print 'uncompressing..'
gz = gzip.open('fpC-'+runstr+'-r'+camcol+'-'+field_str+'.fit.gz')
imgFile = pyfits.open(gz, mode='readonly')
print 'getting header info...'
head = imgFile[0].header
angle = head['SPA']
info = (int(ID), angle)
f.writerow(info)
Tuesday, March 20, 2012
the end of MICE
I've used GALFIT before, while writing my MSc thesis, but oh, those were faint, redshifted and unremarkable SDSS galaxies. That was simple -- I used 5 components once, because a galaxy had a LINER AGN.
The MICE are monsters. They've apparently already passed through each other once, and have highly disturbed tidal features, clumps of active star formation, dust lanes, you name it. I've never used GALFIT to model spiral arms, too, so that was an interesting experience.
Here are they:
I started simple, first masking out each galaxy and fitting the other in turn, then fitting both simultaneously (that's a better way to do it anyway). After running about 150 models I decided to mask out the dark dust lanes in galaxy A -- I couldn't fit them using truncation functions, as GALFIT crashes (on my 64bit SUSE, at least) while attempting that and I don't think the science goal required this. I have some reservations about the decomposition, as I think the image has some flat-fielding issues, besides, the galaxies take up a large part of the overall image, so I have doubts about the determined sky value as well.
In the end I had 91 free parameter, and I think more might have made sense, but that was past the point of diminishing returns. I stopped at Chi^2 = 1.877, and guess the main reason why the model would fit the image so poorly were the bright and irregular cores of the galaxies -- or possible higher order flat-field curvature. The other objects were masked out. Here is the model -- at ds9's 99.5 scale and heat colourmap (this is handy: ds9 -scale mode 99.5 -cmap Heat -medatacube ~/workspace/MICE/galfit/subcomps.fits). The long, bright tails are not well visible with these settings, but they weren't the goal. I am not really happy with the residual picture, as it tells me there are some big, obscured components lurking underneath. But that'll have to do for the moment, as I have other assignments. I've been doing this for other people in the collaboration, so I don't think I can publish the fit logs here, unfortunately.
Friday, March 9, 2012
Healing holes in arrays in Python: interpolation vs. inpainting
I had solved that interpolation task I was working on previously, by the way. I post my approach here, in case someone finds it useful.
If the bad pixel regions are big, standard methods of interpolation are not applicable. Interpolation works under assumption that the regions are contiguous, whereas this is not necessarily the case. Besides, none of interpolation implementations in python seem to work with masked arrays, and I've tried a metric ton of them. miceuz, who was sitting on this task half night as well, asked about it on stackoverflow, and someone mentioned the right keyword: inpainting.That's still inventing the data, but the option is having unquantifiable errors in photometry if we leave out the bad pixels or don't mask the bad areas at all.
OpenCV is a huge piece of software, and using it to fill those holes is tantamount to using a hammer to crack a nut. However, I reused this tiny piece of code, adding a [hardcoded ,)] Gaussian weights kernel as I needed to use inverse distance weighting for interpolation. Here's the code:
import numpy as np
import pyfits
from scipy import ndimage
import inpaint
maskedImg = np.ma.array(cutout, mask = cutoutMask)
NANMask = maskedImg.filled(np.NaN)
badArrays, num_badArrays = sp.ndimage.label(cutoutMask)
print num_badArrays
data_slices = sp.ndimage.find_objects(badArrays)
filled = inpaint.replace_nans(NANMask, 5, 0.5, 2, 'iwd')
hdu = pyfits.PrimaryHDU(filled)
hdu.writeto('filled_lm.fits')
# -*- coding: utf-8 -*-
"""A module for various utilities and helper functions"""
import numpy as np
#cimport numpy as np
#cimport cython
DTYPEf = np.float64
#ctypedef np.float64_t DTYPEf_t
DTYPEi = np.int32
#ctypedef np.int32_t DTYPEi_t
#@cython.boundscheck(False) # turn of bounds-checking for entire function
#@cython.wraparound(False) # turn of bounds-checking for entire function
def replace_nans(array, max_iter, tol,kernel_size=1,method='localmean'):
"""Replace NaN elements in an array using an iterative image inpainting algorithm.
The algorithm is the following:
1) For each element in the input array, replace it by a weighted average
of the neighbouring elements which are not NaN themselves. The weights depends
of the method type. If ``method=localmean`` weight are equal to 1/( (2*kernel_size+1)**2 -1 )
2) Several iterations are needed if there are adjacent NaN elements.
If this is the case, information is "spread" from the edges of the missing
regions iteratively, until the variation is below a certain threshold.
Parameters
----------
array : 2d np.ndarray
an array containing NaN elements that have to be replaced
max_iter : int
the number of iterations
kernel_size : int
the size of the kernel, default is 1
method : str
the method used to replace invalid values. Valid options are
`localmean`, 'idw'.
Returns
-------
filled : 2d np.ndarray
a copy of the input array, where NaN elements have been replaced.
"""
# cdef int i, j, I, J, it, n, k, l
# cdef int n_invalids
filled = np.empty( [array.shape[0], array.shape[1]], dtype=DTYPEf)
kernel = np.empty( (2*kernel_size+1, 2*kernel_size+1), dtype=DTYPEf )
# cdef np.ndarray[np.int_t, ndim=1] inans
# cdef np.ndarray[np.int_t, ndim=1] jnans
# indices where array is NaN
inans, jnans = np.nonzero( np.isnan(array) )
# number of NaN elements
n_nans = len(inans)
# arrays which contain replaced values to check for convergence
replaced_new = np.zeros( n_nans, dtype=DTYPEf)
replaced_old = np.zeros( n_nans, dtype=DTYPEf)
# depending on kernel type, fill kernel array
if method == 'localmean':
print 'kernel_size', kernel_size
for i in range(2*kernel_size+1):
for j in range(2*kernel_size+1):
kernel[i,j] = 1
print kernel, 'kernel'
elif method == 'idw':
kernel = np.array([[0, 0.5, 0.5, 0.5,0],
[0.5,0.75,0.75,0.75,0.5],
[0.5,0.75,1,0.75,0.5],
[0.5,0.75,0.75,0.5,1],
[0, 0.5, 0.5 ,0.5 ,0]])
print kernel, 'kernel'
else:
raise ValueError( 'method not valid. Should be one of `localmean`.')
# fill new array with input elements
for i in range(array.shape[0]):
for j in range(array.shape[1]):
filled[i,j] = array[i,j]
# make several passes
# until we reach convergence
for it in range(max_iter):
print 'iteration', it
# for each NaN element
for k in range(n_nans):
i = inans[k]
j = jnans[k]
# initialize to zero
filled[i,j] = 0.0
n = 0
# loop over the kernel
for I in range(2*kernel_size+1):
for J in range(2*kernel_size+1):
# if we are not out of the boundaries
if i+I-kernel_size < array.shape[0] and i+I-kernel_size >= 0:
if j+J-kernel_size < array.shape[1] and j+J-kernel_size >= 0:
# if the neighbour element is not NaN itself.
if filled[i+I-kernel_size, j+J-kernel_size] == filled[i+I-kernel_size, j+J-kernel_size] :
# do not sum itself
if I-kernel_size != 0 and J-kernel_size != 0:
# convolve kernel with original array
filled[i,j] = filled[i,j] + filled[i+I-kernel_size, j+J-kernel_size]*kernel[I, J]
n = n + 1*kernel[I,J]
print n
# divide value by effective number of added elements
if n != 0:
filled[i,j] = filled[i,j] / n
replaced_new[k] = filled[i,j]
else:
filled[i,j] = np.nan
# check if mean square difference between values of replaced
#elements is below a certain tolerance
print 'tolerance', np.mean( (replaced_new-replaced_old)**2 )
if np.mean( (replaced_new-replaced_old)**2 ) < tol:
break
else:
for l in range(n_nans):
replaced_old[l] = replaced_new[l]
return filled
def sincinterp(image, x, y, kernel_size=3 ):
"""Re-sample an image at intermediate positions between pixels.
This function uses a cardinal interpolation formula which limits
the loss of information in the resampling process. It uses a limited
number of neighbouring pixels.
The new image :math:`im^+` at fractional locations :math:`x` and :math:`y` is computed as:
.. math::
im^+(x,y) = \sum_{i=-\mathtt{kernel\_size}}^{i=\mathtt{kernel\_size}} \sum_{j=-\mathtt{kernel\_size}}^{j=\mathtt{kernel\_size}} \mathtt{image}(i,j) sin[\pi(i-\mathtt{x})] sin[\pi(j-\mathtt{y})] / \pi(i-\mathtt{x}) / \pi(j-\mathtt{y})
Parameters
----------
image : np.ndarray, dtype np.int32
the image array.
x : two dimensions np.ndarray of floats
an array containing fractional pixel row
positions at which to interpolate the image
y : two dimensions np.ndarray of floats
an array containing fractional pixel column
positions at which to interpolate the image
kernel_size : int
interpolation is performed over a ``(2*kernel_size+1)*(2*kernel_size+1)``
submatrix in the neighbourhood of each interpolation point.
Returns
-------
im : np.ndarray, dtype np.float64
the interpolated value of ``image`` at the points specified
by ``x`` and ``y``
"""
# indices
# cdef int i, j, I, J
# the output array
r = np.zeros( [x.shape[0], x.shape[1]], dtype=DTYPEf)
# fast pi
pi = 3.1419
# for each point of the output array
for I in range(x.shape[0]):
for J in range(x.shape[1]):
#loop over all neighbouring grid points
for i in range( int(x[I,J])-kernel_size, int(x[I,J])+kernel_size+1 ):
for j in range( int(y[I,J])-kernel_size, int(y[I,J])+kernel_size+1 ):
# check that we are in the boundaries
if i >= 0 and i <= image.shape[0] and j >= 0 and j <= image.shape[1]:
if (i-x[I,J]) == 0.0 and (j-y[I,J]) == 0.0:
r[I,J] = r[I,J] + image[i,j]
elif (i-x[I,J]) == 0.0:
r[I,J] = r[I,J] + image[i,j] * np.sin( pi*(j-y[I,J]) )/( pi*(j-y[I,J]) )
elif (j-y[I,J]) == 0.0:
r[I,J] = r[I,J] + image[i,j] * np.sin( pi*(i-x[I,J]) )/( pi*(i-x[I,J]) )
else:
r[I,J] = r[I,J] + image[i,j] * np.sin( pi*(i-x[I,J]) )*np.sin( pi*(j-y[I,J]) )/( pi*pi*(i-x[I,J])*(j-y[I,J]))
return r
#cdef extern from "math.h":
# double sin(double)
Thursday, March 8, 2012
Mice and masks
I masked other sources, spurious objects, trails using SeXtractor (it's a pain to compile it from source). However, it's way better to fit only a single galaxy at a time, hence I had to create mask images for them separately.
Doing it the sensible way failed, as the C script would segfault or produce a blank file -- it's probably has to do with ds9 output, but I didn't want to dive into it. Instead, I wrote a simple Python script to mask out rectangular regions around each galaxy.
More elaborate shapes could be made by reusing this script, for instance. When exporting region vertices with ds9, take care to use image coordinates instead of physical, otherwise your mask will look wonky. Here are the Mice galaxies in all their false-colour glory, the image I use for science and one of the final masks.And here goes the essence of that Python script, it's trivial:
maskA = np.zeros((image.shape[0], image.shape[1]))
y = np.zeros(4)
x = np.zeros(4)
for i in range(0, 4):
y[i], x[i] = poly[i]
for i in range(image.shape[0]):
for j in range(image.shape[1]):
if (min(y) < i and i < max(y)) and (min(x) < j and j < max(x)):
maskA[i,j] = 1
Friday, February 24, 2012
Reionisation, healing masked arrays
Two take-home messages were:
- Helium fraction must be included in reionisation simulations
- _If_ (apparently, it's a big if) LOFAR finds a high-redshift radio-loud quasar, they might see the absorbtion features along the line of sight, and obtain better constraints on reionisation parameters.
Later on I was solving a problem that preoccupies me these weeks ahead of the thesis comittee meeting: automated photometry of CALIFA (actually, any SDSS) galaxies, using growth curve analysis for flux extraction. The field stars and other irrelevant objects were already masked by N., but the holes left in the image arrays have to be interpolated, and that's not a trivial task, apparently. Python's interp2d fails on arrays this big (standard SDSS frames, I won't do manual cutouts), besides, I'm not sure it is suitable for this goal, as well as all similar python/numpy routines.
My current approach is finding contiguous bad (=masked) regions in the image array, forming smaller sub-arrays and filling the masked regions within them with the mean of a circular aperture around. That's obviously a naive approach to take -- once I get it running, I'll be able to do more realistic interpolation, e.g. using the galaxy's Sersic index.
It was an interesting detour through numpy array masking, motion detection, image processing and acquainting myself with the slice object. I kind of enjoy diving into those gory details instead of using some nice high level routine -- I'm reinventing the wheel, obviously, but that's more scientific than using black box methods.
Finding contiguous regions within an array, apparently, is not that trivial as I first expected, but the solution I used was really helpful -- stackoverflow rocks as usual.